CATEGORY
Filter
SOURCE
Squonk
DESCRIPTION
De-duplicate a set of structures using a canonical smiles field
INPUTS
A Dataset of Molecules (must already contain the canonical smiles field).
OUTPUTS
A Dataset of Molecules, with the structures being the canonical smiles.
OPTIONS
| Canonical smiles field | The name of the field containing the canonical smiles |
| Keep first value fields | When a duplicate is identified keep the first value that is encountered for these fields |
| Keep last value fields | When a duplicate is identified keep the last value that is encountered for these fields |
| Append all values fields | When a duplicate is identified append all values to a list for these fields. The result for those fields is always a list, even if not duplicates were encountered |
When specifying multiple fields separate the names with commas. Note: a UI that allows the fields to be selected from a list is planned.
ADDITIONAL INFO
The screenshot below is an example of this cell in use. Structures are read from a SD file, canonical smiles is generated and then de-duplicated using this cell, in this case generating a list of encountered values for three fields.
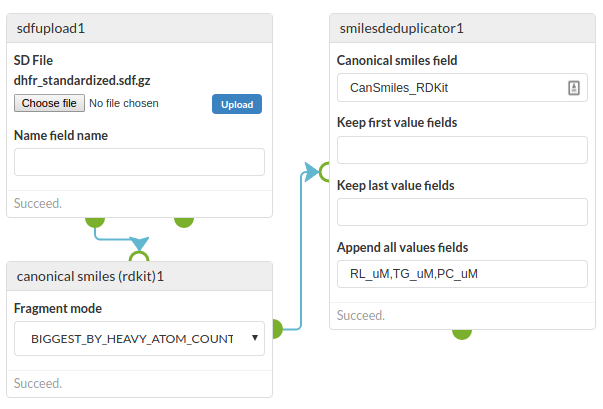
De-duplication is a common need when you have a set of structures. The structures may be defined in different ways (e.g. aromatic and Kekule forms) and may have salts which you may want to ignore. Whilst there are various approaches to de-duplication a simple, but fairly robust, one is to generate a canonical smiles string for each structure sand use those to determine the equivalence of the structures. Depending on how the canonical smiles is generated you can determine how molecules with multiple fragments (e.g. salts) are handled.
The success is dependent on the quality of the canonicalisation algorithm and other factors such as structure standardization/sanitization, so we do not include a specific algorithm in this cell, instead requiring this to be already present in the input (usually this means generated by an upstream cell). We expect to have a number of such canonical smiles generators. Here is a (not necessarily complete) list of the current ones:
A key thing when de-duplicating is what to do with the properties of the duplicate molecules. Currently this cell has a few simple ways to handle this (see the options above) though we plan to add others (such as min and max value) in future.