CATEGORY
Clustering
SOURCE
RDKit
DESCRIPTION
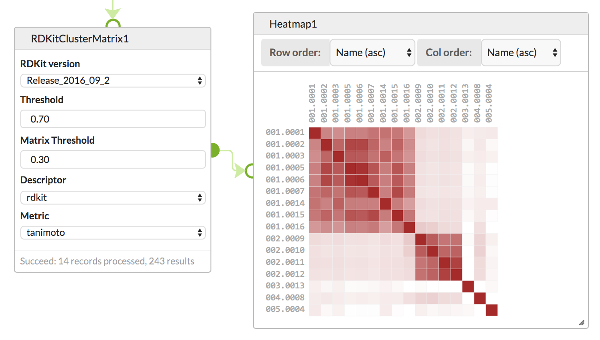
Generates a clustered distance matrix from a dataset of molecules using Butina clustering from RDKit. Is usefully combined with the Heatmap cell for displaying the distance matrix
INPUTS
A dataset of molecules
OUTPUTS
A dataset of basic objects containing the similarity scores for each cell in the matrix
OPTIONS
| Threshold | The similarity threshold for clustering |
| Matrix threshold | The similarity threshold for including the comparison in the output. If the similarity is less than the specified value it is excluded. Useful for reducing the size of the output by removing low similarities |
| Descriptor | Fingerprint type. Options are: maccs, morgan2, morgan3, rdkit (default |
| Metric | Similarity comparison metric. Options are: asymmetric, braunblanquet, cosine, dice, kulczynski, mcconnaughey, rogotgoldberg, russel, sokal, tanimoto (default) |
ADDITIONAL INFO
The cell outputs a similarity matrix for a set of structures, and includes IDs that reflect the cluster that structure corresponds to. The Similarity threshold, Descriptor and metric determines the clustering. The Matrix threshold determines which scores are output.
A common use for this cell is to display the similarities amongst a set of structures using the Heatmap cell.